Geoduck larvae low pH trials
05/09/17
We will be conducting another geoduck larvae experiment at the Taylor Shellfish Hatchery and sampling for proteomics and microbiomes. Since late February we have conducted about 10 spawns and fertilized in total about 600 million eggs. Out of these 600 million eggs we have only managed to get out about 3 million competent eyed larvae. Mortality is high and we see ciliate/ameoba infestations occuring anywhere from Day 5 to Day 13. Like with our oyster experiments, we are unsure whether the ciliates/ameobas are invasive (predatory) and infecting healthy larvae or if they are scavengers feeding on dying tissue. We will use proteomics to reveal any potential underlying physiological stressors that may be impacting these larvae during these early stages of life. Additionally, we will filter effluent seawater from the conicals to look at the microbial communties that may be associated with high mortality events.
In 2016, we reported greater survival to setting when geoducks were transferred from pH 8.2 to pH 7.1 at Day 4. In this experiment we will try a similar approach. We will hatch all eggs in deep seawater which is pumped in from Dabob Bay at 100 feet deep and treated with sodium carbonate to pH 8.2. At day 4 we will split the larval group in half- one half staying at pH 8.2 and one half transferred to a low pH group at pH 7.1. This low pH group is deep raw seawater pumped in from Dabob Bay but untreated with sodium carbonate. Instead this water is bubbled with CO2 to reach pH 7.1. All seawater used for the experiment is filtered through 25, 10, and 5 um bag filters and heated to 14C. I have placed Hobo Light/Temperature sensors in each conical to monitor temperature fluctuations as this experiment is occuring in a greenhouse.
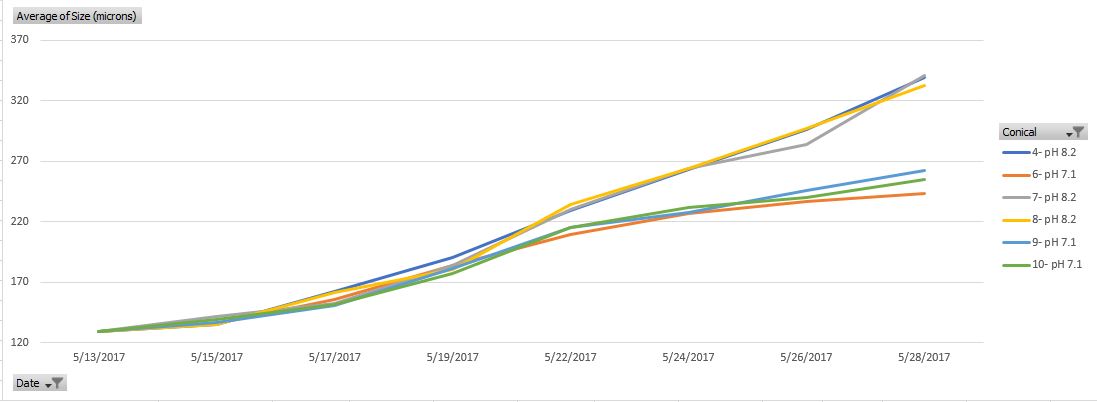
Morphology
- Prior to tank change that will occur on a Monday-Wednesday-Friday schedule I will sample ~100 larvae from each conical using a small mesh screen and pipette. Using a compound microscope I will measure 50 larvae and document whether alive or dead. Using this information I will be able to determine percent mortality prior to tank change and know what screen I can use to remove sick and dead larvae.
Proteomics
- Following morphology samples, I will again use a small mesh screen to capture larvae from each conical by scooping. I will give a genetle rinse using distilled water and quickly pipette out larvae with the least amount of water possible to transfer larvae to a cryotube. I will flash freeze before storing at -80C.
Microbiomes
- For microbiome sampling I will fill a gallon container with effluent seawater from each conical. I will use a peristaltic pump to filter the seater through three filters using Swinnex filter holders. The three filters are 5um, 0.7um, and finally a 0.22um to retain bacteria. I will save all three filters from each conical and store at -80C. I will do this every other sampling day.
Schedule:
5/15/17
5/19/17
5/24/17
5/29/17
6/2/17
Quantification
- To quantify larvae I will drop the conical and collect all larvae in a 3L pitcher. I will plunge the sample to distribute the larvae evenly within the water column. Based on the previous quantification I will calculate an approximate volume I will need to pipette in order to count at least 100 larvae. I will repeat this three times to get 3 counts within 10% of eachother. Then I can calculate how many larvae I have within each conical. This will also be done on every other sampling day- alternating with microbiome sampling.
Schedule:
5/17/17
5/22/17
5/26/17
5/31/17
6/5/17
UPDATES:
5/8/17
I did a little practice run with the hobo loggers to make sure they would all read the same temperature when held in the same tank.
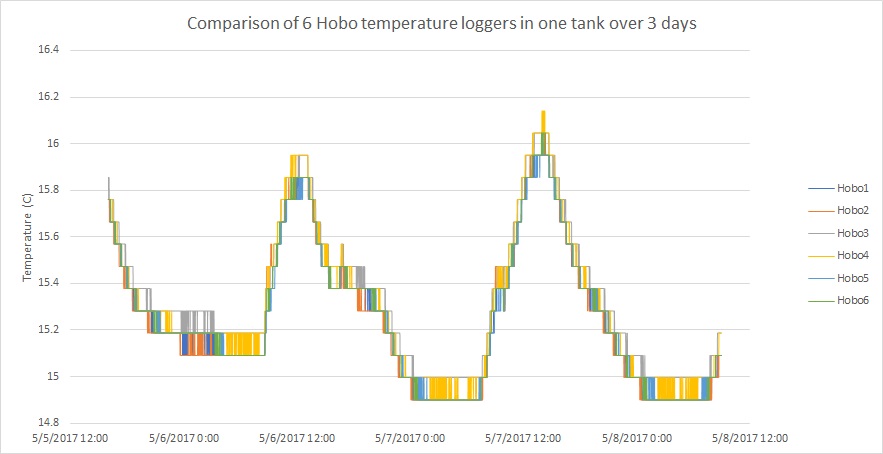
Looks pretty good!
5/9/17
We induced a natural geoduck spawn by raising seawater temperature from 14C to 16C and giving the animals higher algae rations. We also drain and refill the totes several times to initiate spawning.
Sucess!! We pooled the eggs from several females and screened using an 80um to remove other biological material and a 20um screen to retain eggs. We divided the pooled eggs into the number of males we got. Then we fertlized each bucket of eggs with a different male. In total, we have 30 millions eggs to hatch in our research system in the new greenhouse! I divided the eggs up evenly in 5 conicals with 28um banjos. I put a Hobo logger in each one.
5/11/17
At 8AM in the morning, I checked conicals for D-hinge larvae. There were still many trochophores swimming around so I gave each conical 3L of food and gave them until the end of the day to develop. At 3PM I dropped all of the conicals on 48 um screens and combined into one screen. I checked to see if the larvae held on a 60 um screen and most did so I dumped the remaining colorless/underdeveloped larvae. I used a coulter counter to count the number of D-hinge I had in 5L. The coulter counter displays readings in # of larvae per ml.
2474, 2358, 2466 Average = 2433 *5000 = 12,163,333 larvae
I divided this evenly amoung 5 conicals all at pH 8.2. I started feeding with incoming microalgae densities of ~50,000 cells/ml.
5/13/17
First thing in the morning, I sampled larvae for proteomics. Because I want to sample proteomics before I stress the larvae out in anyway (by screening) I needed to take it from a random conical- it happened to be Conical 7. I also took a morphology sample and put it in the fridge to analyze in a few hours.
I dropped the conicals on 60 um screens and collected all larvae into one screen. I set aside while I chlorinated the whole system and began the treatments of low and high pH.
Conical Layout
- 4: pH 8.2
- 6: pH 7.1
- 7: pH 8.2
- 8: pH 8.2
- 9: pH 7.1
- 10: pH 7.1
All larvae were pooled in a bucket, filled to 6L and plunged to redistribute 1L into each of the six conicals. Each conical has a 60um banjo.
All of the larvae were distributed into their new homes.
5/15/17
Here are some photos to start off today’s post:
Filtration set up as presented by Justin Thompson
My pH7.1 header which is seawater bubbled with CO2
My pH8.2 header which is seawater treated with sodium carbonate
Conicals
Conical with pH 7.1
Conical with pH 8.2
Microbiome sampling
Today was the first full sampling day of all conicals.
I started off the day by sampling each conical for proteomics. Then I took larval samples for microscopy. While I was measuring larvae Justin started on the seawater filtering. We collected the effluent from the conical within 30 minutes of filtration. We filtered seawater from 9AM until about 4PM. The middle filter 0.7um kept tearing which meant we had to redo the whole process.
After the effluent was filtered from a conical I dropped it and started screening larvae. I dropped each conical on a 60um and decided to keep 80s and above because the larvae that fell through the 80um looked abnormally developed/unhealthy.
I am seeing some abnormal velums and swimming behavior in both treatments.
pH 8.2 larvae
pH 7.1 larvae
5/17/17
Today I am going to quantify the larvae in each conical. Before doing counts however, I will sample for proteomics and morphology. I am guessing I have about 1- 1.5 million eggs so I calculated the volume required to count about 100 oysters from a 3L pitcher. If I pipette 200ul aliquots, I should be able to count an adequate number of larvae to estimate what I have in each conical. I will start with three replicates but if I need to do more I can.
Conical 4: pH 8.2
Count 1: 69
Count 2: 68
Count 3: 72
Average: 69.67 *3000ml / 0.2ml = 1,045,000
Number of larvae in 3L pitcher: 1,045,000
Conical 6: pH 7.1
Count 1: 76
Count 2: 56
Count 3: 84
Count 4: 82
Count 5: 96
Count 6: 87
Average: 80.17 *3000ml / 0.2ml = 1,202,500
Number of larvae in 3L pitcher: 1,202,500
Conical 7: pH 8.2
Count 1: 75
Count 2: 50
Count 3: 59
Count 4: 64
Count 5: 64
Count 6: 60
Average: 62.00 *3000ml / 0.2ml = 930,000
Number of larvae in 3L pitcher: 930,000
Conical 8: pH 8.2
Count 1: 75
Count 2: 58
Count 3: 75
Count 4: 69
Count 5: 76
Count 6: 85
Average: 73.00 *3000ml / 0.2ml = 1,095,000
Number of larvae in 3L pitcher: 1,095,000
Conical 9: pH 7.1
Count 1: 77
Count 2: 65
Count 3: 83
Count 4: 85
Count 5: 86
Count 6: 71
Average: 77.83 *3000ml / 0.2ml = 1,167,500
Number of larvae in 3L pitcher: 1,167,500
Conical 10: pH 7.1
Count 1: 75
Count 2: 55
Count 3: 72
Count 4: 46
Count 5: 62
Count 6: 63
Average: 62.17 *3000ml / 0.2ml = 932,500
Number of larvae in 3L pitcher: 932,500
5/19/17
Brent helped out with microbiome sampling today. No quantification today but did proteomics and morphology. I kept all larvae holding on 100 micron screen.
5/22/17
Grace came out today and helped with microbiome sampling. I determined the number of larvae per conical by taking 300ul subsamples from a well distributed 3L pitcher of larvae. Here are my counts:
Conical 4: pH 8.2
Count 1: 84
Count 2: 61
Count 3: 69
Count 4: 67
Count 5: 65
Count 6: 57
Average: 67.17 *3000ml / 0.3ml = 671,667
Number of larvae in 3L pitcher: 671,667
Conical 6: pH 7.1
Count 1: 76
Count 2: 71
Count 3: 59
Count 4: 60
Count 5: 83
Count 6: 84
Average: 72.17 *3000ml / 0.3ml = 721,667
Number of larvae in 3L pitcher: 721,667
Conical 7: pH 8.2
Count 1: 56
Count 2: 54
Count 3: 52
Count 4: 61
Count 5: 51
Count 6: 61
Average: 55.83 *3000ml / 0.3ml = 558,333
Number of larvae in 3L pitcher: 558,333
Conical 8: pH 8.2
Count 1: 37
Count 2: 73
Count 3: 85
Count 4: 71
Count 5: 54
Count 6: 67
Average: 64.50 *3000ml / 0.3ml = 645,000
Number of larvae in 3L pitcher: 645,000
Conical 9: pH 7.1
Count 1: 81
Count 2: 70
Count 3: 81
Count 4: 41
Count 5: 59
Count 6: 82
Average: 69.00 *3000ml / 0.3ml = 690,000
Number of larvae in 3L pitcher: 690,000
Conical 10: pH 7.1
Count 1: 88
Count 2: 70
Count 3: 60
Count 4: 60
Count 5: 32
Count 6: 83
Average: 65.50 *3000ml / 0.3ml = 655,000
Number of larvae in 3L pitcher: 655,000
After quantification, I kept all larvae holding on a 118um screen.
5/24/17
No microbiome sampling or quantification today. Just morphology and proteomics. I kept all larvae holding on a 132um screen.
5/26/17
Quantification, morphology, and proteomics. Grace came out to microbiome sample.
Conical 4: pH 8.2
Count 1: 70
Count 2: 58
Count 3: 62
Count 4: 62
Count 5: 52
Count 6: 71
Average: 62.50 *3000ml / 0.3ml = 625,000
Number of larvae in 3L pitcher: 625,000
Conical 6: pH 7.1
Count 1: 69
Count 2: 65
Count 3: 49
Count 4: 44
Count 5: 55
Count 6: 52
Average: 55.67 *3000ml / 0.3ml = 556,667
Number of larvae in 3L pitcher: 556,667
Conical 7: pH 8.2
Count 1: 27
Count 2: 54
Count 3: 42
Count 4: 33
Count 5: 38
Count 6: 38
Average: 38.67 *3000ml / 0.3ml = 386,667
Number of larvae in 3L pitcher: 386,667
Conical 8: pH 8.2
Count 1: 68
Count 2: 49
Count 3: 37
Count 4: 49
Count 5: 44
Count 6: 58
Average: 50.83 *3000ml / 0.3ml = 508,333
Number of larvae in 3L pitcher: 508,333
Conical 9: pH 7.1
Count 1: 79
Count 2: 76
Count 3: 82
Count 4: 79
Count 5: 89
Count 6: 71
Average: 79.33 *3000ml / 0.3ml = 793,333
Number of larvae in 3L pitcher: 793,333
Conical 10: pH 7.1
Count 1: 61
Count 2: 58
Count 3: 61
Count 4: 70
Count 5: 62
Count 6: 76
Average: 64.67 *3000ml / 0.3ml = 646,667
Number of larvae in 3L pitcher: 646,667
5/28/17
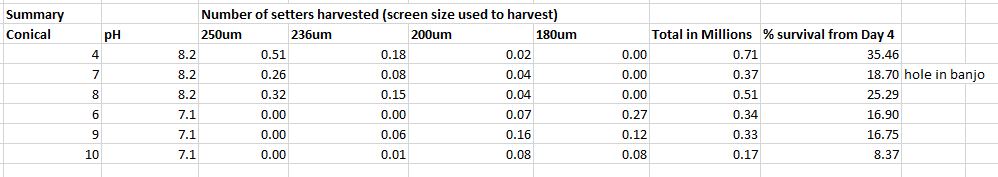
Sampled for morphology and proteomics- last time. I am harvesting setters today.
pH 8.2 Treatment
Conical 4: 175K- 250s
Conical 7: 75K- 250s
Conical 8: 105K- 250s
These pH 8.2 setters were combined and set in 4 silos.
No setters in pH 7.1 treatment
5/30/17
Harvested Setters
pH 8.2 Treatment
Conical 4: 335K- 250s and 167K- 236s
Conical 7: 180K- 250s and 78K- 236s
Conical 8: 215K- 250s and 139K- 236s
No setters in pH 7.1 treatment
6/1/17
Harvested setters and end of experiment
pH 8.2 Treatment
Conical 4: 11K- 250s and 21K 236s
Conical 7: 6K-250s and 36K 236s
Conical 8: 11K- 250s and 36K 236s
pH 7.1 Treatment
Conical 6: 71K- 200s and 267K -180s
Conical 9: 56K- 236s and 157K 200s and 122K 180s
Conical 10: 11K 236s and 79K 200s and 78K 180s
Dumped all remaining larvae as it looked pretty bad. End of experiment.